More Information
Submitted: February 14, 2023 | Approved: March 03, 2023 | Published: March 04, 2023
How to cite this article: Deng X, Li H, Qiao J, Tong Y, Wang C, et al. The effects of exercise intensity on the gut microbiota of college basketball players. J Community Med Health Solut. 2023; 4: 010-018.
DOI: 10.29328/journal.jcmhs.1001028
Copyright License: © 2023 Deng X, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Athletes; Exercise intensity; Gut microbiota; High-throughput sequencing
Acronyms and Abbreviations: MHR: Maximum Heart Rate; L: Low-Intensity Exercise Group; M: Medium-Intensity Exercise; H: High-Intensity Exercise
The effects of exercise intensity on the gut microbiota of college basketball players
Hong-xian Deng1, Huan-huan Li1, Jiu-quan Qiao2*, Yan Tong1*, Cui-juan Wang1 and Jiang Liu2
1School of Life Science and Engineering, Southwest Jiaotong University, Chengdu, China
2School of Physical Education, Southwest Jiaotong University, Chengdu, China
*Address for Correspondence: Yan Tong, School of Life Science and Engineering, Southwest Jiaotong University, Chengdu, China,
Email: [email protected]
Jiu-quan Qiao, School of Physical Education, Southwest Jiaotong University, Chengdu, China, Email: [email protected]
Purpose: Exercise has a positive regulatory effect on gut microbiota and is also involved in regulating multiple physiological functions of the human body. This article tested the effects of basketball exercises at different exercise intensities on the gut microbiota of college students.
Methods: Athlete research subjects (male, aged 18 - 25) were selected from the basketball team and trained at different intensities to obtain a total of 101 fresh fecal samples. DNA was extracted by a DNA extraction kit and bacterial 16S rRNA gene V3-V4 region high-throughput sequencing using the Illumina Hiseq platform. The downstream data were spliced, filtered and de-trimerized and then used to study the difference in gut microbiota.
Results: Key bacterial taxa in the gut that responded to exercise intensity differed among athletes of different exercise intensities but most belonged to Firmicutes. With increasing exercise intensity, Butyricicoccus, Anaerostipes, Oxalobacter and Clostridium_IV in basketball players enrich. Further analysis of the functional prediction revealed that carbohydrate metabolism, amino acid metabolism, metabolism of xenobiotics and glycans and metabolism were significantly expressed in the gut microbiota of basketball athletes with high intensity.
Conclusion: The study demonstrated that after long-term professional training, the gut microbiota of athletes adapts to exercise stimulators and can quickly respond to changes in exercise intensity. In high-intensity training, the organism is protected from harm by enriching some beneficial bacteria.
As a healthy lifestyle, regular physical activity and moderate exercise can not only effectively improve human health and quality of life, but also can enhance the body’s anti-inflammatory capacity by optimizing physiological and neuroendocrine stress responses [1]. The human gastrointestinal tract (GI) contains a rich and diverse microbial community of over one trillion microorganisms that interact with each other and their hosts [2]. The gut microbiome is associated with a range of human diseases [3]. Although the evidence for many of these diseases is not strong, it has long been hypothesized that gut microbiota has an important role in maintaining normal gut function and the overall health of individuals. Research on the correlation between gut flora function and exercise is still in its infancy. Increasing evidence points out that exercise had a positive regulatory effect on gut microbiota [4]: regulating effectively the composition and the structure of gut microbiota, improving the richness of gut microbiota, increasing the number of beneficial gut microbiota, maintaining the balance of flora and thus improve body health. Alex E Mohr, et al. [5] found that exercise can promote healthy bacteria with rich species, increased microbial diversity, functional metabolic capacity, and microbial-related metabolites. Stimulating bacterial abundance can regulate mucosal immunity and improve gastrointestinal barrier function. Sabrina Donati Zeppa [6] found that exercise has been proven to increase the diversity of microbiota, thereby improving metabolic characteristics and immune response, and also regulate the energy balance and participate in the control of the influence of individual intestinal microbiota in the state of inflammation, redox and hydration. David M Keohane, et al. [7] found that long-term and high-intensity exercise positively affected intestinal microbial diversity, increased the relative abundance of some bacterial species and up-regulated the metabolic potential of specific pathways expressing microbial gene products.
Gut microbiota is involved in the regulation of several physiological functions in the human body and influences the development of diseases. They can encode a wide variety of enzymes and are closely related to the nutritional, metabolic and immune aspects of the host, and are influenced by various factors such as genetic background, age, health status, dietary habits, drug use, and lifestyle of the host [8]. Most of the research has been conducted on rodents. Preliminary studies have shown that exercise not only changes the structure of the gut microbiota in healthy animals but also promotes the metabolic function of the gut microbiota and influences the functional activity of the “Microbiota-Gut-Brain Axis” [9]. In addition to the above data obtained through animal studies, the same results were obtained in population studies, suggesting that exercise improves the structure of the gut microbiota in the human body. Clarke, et al. [10] found that professional rugby players had significantly different gut microbiota than healthy non-athlete individuals, with the former having a higher gut microbiota diversity and abundance and that exercise reduced the inflammatory response in athletes.
To date, there have been relatively few systematic studies on the effects of specific exercise forms, exercise duration, exercise intensity and exercise volume on gut microbiota [11]. At present, in China, the relationship between exercise and intestinal microflora is not perfect for the guidance of non-professional sports teams. However, due to the poor lifestyle and eating habits in modern life, the frequency of obesity and intestinal inflammation in modern people has increased significantly. Based on the 16S rRNA gene high-throughput sequencing technology, the composition, and structure of fecal microflora of male basketball team athletes at different intensities of exercise were analyzed. From the perspective of intestinal microflora, the effects of different exercise intensities on human intestinal function were studied, and the main bacterial groups responding to changes in exercise intensity were identified, which made us understand the response of the human gastrointestinal tract to different intensities of exercise stimuli from a new perspective. Analyze the intestinal community structure of non-professional athletes, reveal the principle of action of sports on the intestinal flora and give some sports suggestions to the general public, so as to make suitable sports guidance for people at all stages of health.
Subject characteristics
A total of 27 professional basketball athletes (all males) were recruited from the undergraduate students at Southwest Jiaotong University, China. In this study, students from 18 years - 25 years of age who joined the school basketball team at the same time were selected for the experiment. The standards of physical exercises are shown in Table 1. Exclusion criteria: gastrointestinal diseases; antibiotic treatment within the previous three months; autoimmune diseases; interventional surgical treatment. In the early stage of the experiment, we conducted a one-month diet, physical activity, and lifestyle control for basketball team members. The dietary variety of the athletes remained relatively fixed during the trial period (No probiotics, coffee, alcohol, etc). Each of them signed a written consent for the acknowledgment and approval of applying their data and samples for scientific research purposes. Ethical approval was given by the Ethics Committee of Southwest Jiaotong University. The study conforms to the principles of the Helsinki Declaration.
| Table 1: Standards of physical exercise of basketball athletes. | ||||
| Groups | Exercise intensity | Exercise time | ||
| CK | Before exercise | —— | —— | —— |
| L | Low-intensity exercise | 60% - 70% MHR | recovery of physical function | training on Monday, Wednesday, and Friday, 2 h each time for 3 weeks |
| M | Medium-intensity exercise | 70% - 80% MHR | two peaks per training session | |
| H | High-intensity exercise | 80% - 90% MHR | two peaks per training session | |
Fecal sample collection, DNA extraction and PCR
Fecal were collected the day after the athlete completed the phasic training and loaded in the sterile collection containers, which were stored at - 80 °C until extraction. DNA was extracted and purified using QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). The integrity and concentration of genomic DNA were respectively tested by agarose gel electrophoresis and NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA).
Toptaq DNA polymerase Kit (transgen, China) was used to amplify the qualified samples in ABI 2720 Thermal Cycler (Thermo Fisher Scientific, USA) and the standard bacterial genomic DNA Mix was used as a positive control in three replicate reactions. We used the bacterial genomic DNA as the template to amplify the V3 - V4 hypervariable region of the 16S rRNA gene, and the amplification primer was determined according to the selected detection region, the specific sequence is as follows: primer F (Illumina adapter sequence 1+ CCTACGGGNGGCWGCAG) and primer R (Illumina adapter sequence 2+ GACTACHVGGGTATCTAATCC), Illumina adapter sequence 1 (CTGTCTCTTA TACACA TCTCCGAGCCCACGAGAC) and Illumina adapter sequence 2 (CTGTCTCTTA TACACA TCTGACGCTGCCGACGA). The singleness and specificity of the amplified products were detected by agarose gel electrophoresis.
Microbial sequencing and sequence quality control
PCR products were sequenced using the Illumina Miseq platform with the 2 × 250 bp paired-end sequencing method by Genesky Biotechnologies Inc. (Shanghai, China) after the library was quantified, mixed, and quality checked [12]. Raw reads were quality filtered and merged by several steps: (1) Raw reads and adapter sequences at average quality score < 20 were filtered with TrimGalore and further removal of reads with length < 100 bp; (2) pairs of reads from the original DNA fragments were merged using FLASH2 [13], and low-quality sequences were further removed after merging (More than 90% of base quality is less than 20); (3) Mothur [12] was used to find and remove the primers in the sequence, as well as to delete sequences including n-bases / homopolymers > 6 bp; (4) Reads with an error rate > 2 and reads with length < 100 bp were removed using USEARCH to obtain clean reads for further analysis. Operational Taxonomic Units (OTUs) by UPARSE after clustering sequences and removal of chimeras at 97% similarity [14].
Community diversity analysis
OTU clustering was performed at a threshold of 97% sequence similarity to get the number of OTU per sample. Venn diagram was used to demonstrate the number of shared and respective unique OTUs in multiple samples and to visually show the overlap of OTUs among samples. The PCoA analysis is based on the Bray-Curtis distance for β diversity analysis by R scatterplot3d. Lefse analysis identifies m intestinal microbiomes, especially biomarkers. Different abundant bacterial taxa among the groups were identified using the Linear Discriminant Analysis (LDA) effect size (LEfSe) [15]. Only those taxa that obtained a log-LDA score > 2 were ultimately considered. KEGG pathways functions analysis was performed with PICRUSt based on 16S rRNA sequencing data.
Statistical method
The data were expressed as means ± standard deviation. SPSS version 26.0 (IBM, Chicago, USA) was used to perform one-way ANOVA to analyze the significance of differences among data in each group and the t-test test to analyze the difference between the two groups. Mann-Whitney test was performed with R to calculate the difference of OTU relative abundance. A two-sided p < 0.05 was considered to achieve statistical significance.
Participant characteristics
As shown in Table 2, the participants’ characteristics did not differ between any of the groups.
| Table 2: Characteristics of subjects in the study. | ||||
| Characteristics | Groups | |||
| CK (n = 26) | L (n = 27) | M (n = 25) | H (n = 23) | |
| Age (years) | 19.76 ± 0.89 | 19.51 ± 0.91 | 19.64 ± 0.97 | 19.65 ± 0.86 |
| Height (cm) | 182.65 ± 6.46 | 181.81 ± 6.26 | 181.8 ± 6.19 | 183.52 ± 6.28 |
| Weight (kg) | 78.64 ± 11.84 | 76.6 ± 11.34 | 76.97 ± 9.25 | 80.35 ± 11.74 |
| BMI (kg/m2) | 23.57 ± 0.28 | 23.17 ± 0.29 | 23.28 ± 0.24 | 23.85 ± 0.30 |
| Diet | Equal protein, fat and carbs | |||
| Resting heart rate (bpm) | 66.92 ± 7.95 | 66.4 ± 7.72 | 66.44 ± 8.34 | 65.6 ± 7.73 |
| Maximum heart rate (bpm) | 108.23 ± 0.24 | 130.29 ± 0.54 | 150.24 ± 0.66 | 170.29 ± 0.73 |
Sequencing results, OTU clustering and species annotation
The number of input sequences was 8,268,869 ± 2990.74, and a total of 6,824,841 ± 4672.57 high-quality sequences were detected after de-chimerism among the 101 fecal samples from the participants with an average of 67,572 ± 46.26 sequences per sample. The distribution of clean sequences clustered at length 425 bp. A total of 644 species classifications were obtained from 101 samples. The fecal bacteria were composed of 18 phylum, 31 classes, 44 orders, 93 families, 226 genus and 232 species in total. Compared with the CK group, the low-intensity group had a bigger number of bacterial taxa at each level (species, genus, family, order, class and phylum) (Table 3).
| Table 3: Bacterial compositions in each group at different levels. | ||||||
| Taxon | Total | Overlap | CK | L | M | H |
| phylum | 18 | 10 | 3 | 3 | 6 | 2 |
| class | 31 | 15 | 6 | 9 | 13 | 2 |
| order | 44 | 21 | 10 | 11 | 11 | 3 |
| family | 93 | 48 | 14 | 27 | 20 | 8 |
| genus | 226 | 113 | 34 | 72 | 33 | 25 |
| species | 232 | 78 | 85 | 85 | 45 | 56 |
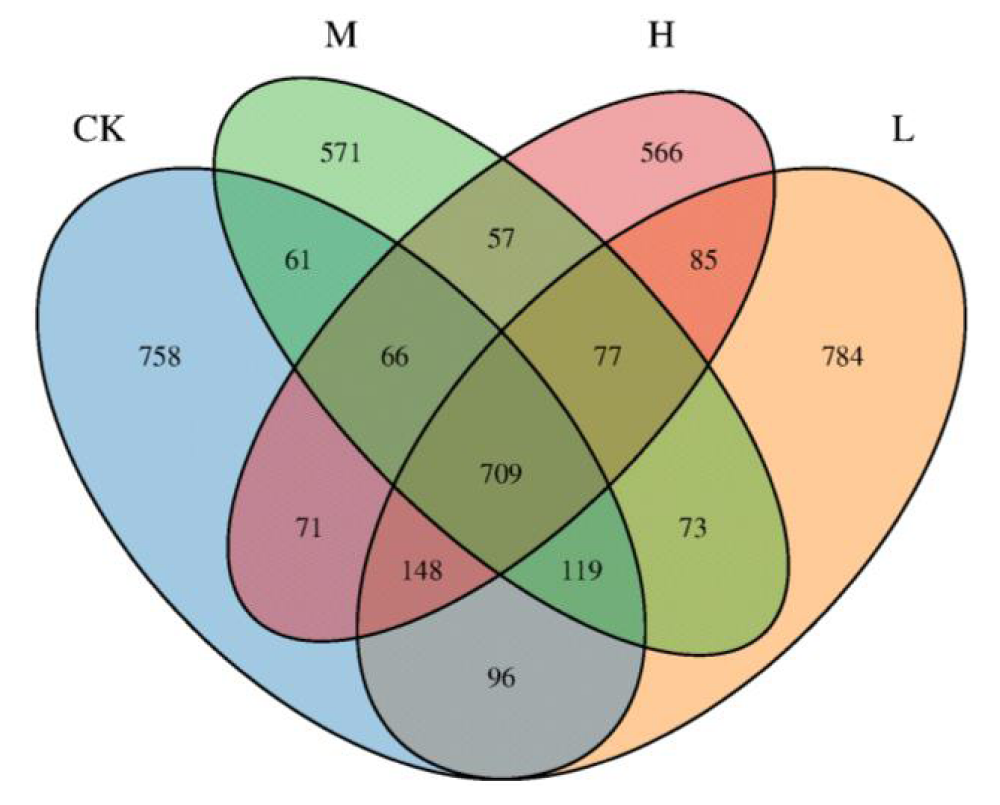
A total of 969 OTUs were quantified in this study. The Venn diagram showed that the number of OTUs was bigger in the CK group (784) than in the other three groups (CK: 758, M: 571, H: 566). There are 709 OTUs with highly similar gut microflora in the four groups of athletes with 16.72% of the total OTUS, and the proportions of each group were as follows: CK (18.87%), L (18.49%), M (13.46%), H (13.35 %) (Figure 1).
Figure 1: Venn diagram of OTU distribution of gut microbiota in four groups of athletes.
Sequencing depth analysis
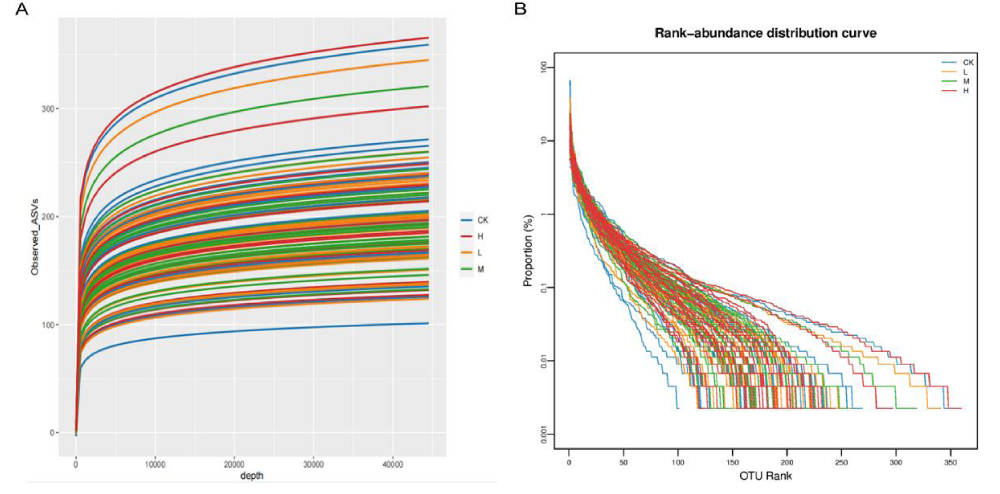
The flat trend of the rarefaction curve indicates that large enough sequencing depth and data volume for data analysis in this experiment. As the sequencing depth increased, the rarefaction curve of CK, L, M and H gradually leveled off with a decreasing slope when the sample depth reached 44000. Thus, the sequencing depth of this study was sufficient to meet the requirements of the subsequent analysis (Figure 2A).
The richness and evenness of species in each group were investigated by rank-abundance curve (Figure 2B). Compared with the CK, the horizontal width of the curve was wider and flatter in shape in M and H, indicating a higher abundance and even distribution of species in the gut microbiota of groups M and H.
Figure 2: Sequencing depth of gut microbiota for basketball athletes. A: Rarefaction curve of gut microbiota in four groups; B: Rank-abundance distribution curve of gut microbiota in four groups.
α-diversity
The community richness index (Observed, Chao1, ACE) and the community diversity index (Shannon, Simpson, Coverage) between the two groups increased with the exercise intensity. However, the correlation was not significantly different between groups with Kruskal-Wallis analysis (Table 4), indicating that different exercise intensities of basketball players have no significant effect on the community richness and the diversity of their body flora during long-term exercise.
| Table 4: Effect of exercise intensity on the diversity of gut microbiota in athletes. | |||||||
| Groups | n | Observed | Chao1 | ACE | Shannon | Simpson | Coverage |
| CK | 26 | 191.76 ± 40.66 | 195.49 ± 41.85 | 195.76 ± 42.07 | 3.74 ± 0.57 | 0.06 ± 0.08 | 1 ± 0 |
| L | 27 | 199.33 ± 46.8 | 203.5 ± 54.33 | 203.6 ± 48.34 | 3.73 ± 0.37 | 0.05 ± 0.03 | 0.99 ± 0 |
| M | 25 | 200.26 ± 51.58 | 203.5 ± 48.43 | 203.77 ± 52.79 | 3.84 ± 0.33 | 0.04 ± 0.01 | 0.99 ± 0 |
| H | 23 | 200.46 ± 53.74 | 203.34 ± 52.73 | 204.1 ± 54.39 | 3.89 ± 0.35 | 0.04 ± 0.01 | 0.99 ± 0 |
Species composition of difference analysis
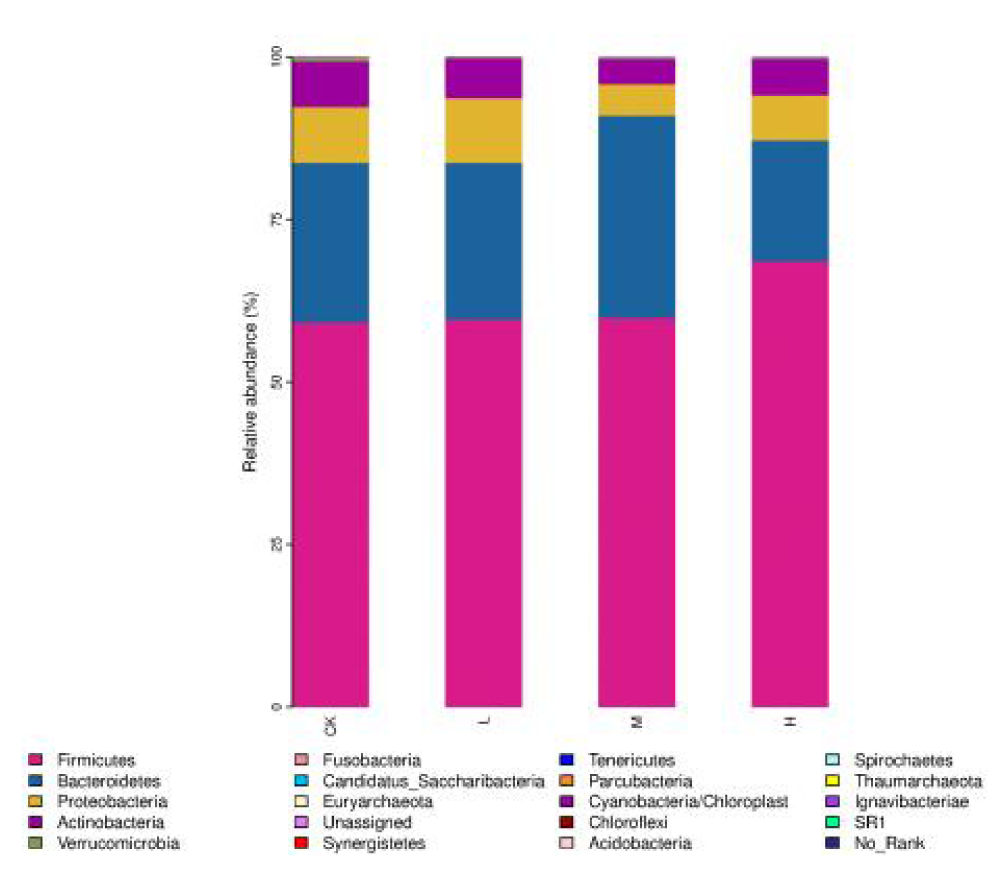
The gut microbiota is dominated at the phylum level by the Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Figure 3). Compared with the CK, the abundance of Bacteroidetes in the gut microbiota increased in the M and decreased in the H. However, Parcubacteria increased in the H. At the genus level, nine genera were significantly increased in relative abundance in M and H compared to CK, including Butyricicoccus, Anaerostipes, Oxalobacter and Clostridium_IV (Table 5).
Figure 3: Effect of exercise intensity on the composition of gut microbiota of basketball athletes at the phylum level.
| Table 5: Comparison of differential species of gut microbiota before and after exercise. | |||||||
| Taxon name | Relative abundance (%) | p - Value | FDR q Value | ||||
| CK | L | M | H | ||||
| Phylum | Parcubacteria | < 0.001 | < 0.001 | < 0.001 | 0.002 | 0.009 | 0.182 |
| Bacteroidetes | 24.513 | 24.110 | 31.129 | 18.616 | 0.0437 | 0.395 | |
| Genus | Pseudomonas | 0.845 | 0.477 | 0.001 | < 0.001 | < 0.001 | < 0.001 |
| Stenotrophomonas | 0.031 | 0.022 | 0.003 | < 0.001 | < 0.001 | < 0.001 | |
| Gemella | 0.0256 | 0.017 | 0.002 | 0.021 | < 0.001 | 0.049 | |
| Ruminococcus2 | 3.934 | 2.797 | 1.815 | 1.696 | 0.003 | 0.153 | |
| Butyricicoccus | 0.473 | 0.7033 | 0.937 | 1.438 | 0.012 | 0.234 | |
| Anaerostipes | 0.640 | 1.011 | 1.423 | 1.575 | 0.014 | 0.395 | |
| Oxalobacter | < 0.001 | 0.002 | 0.003 | 0.010 | 0.049 | 0.671 | |
| Clostridium_IV | 0.098 | 0.202 | 0.308 | 0.348 | 0.049 | 0.671 | |
Biomarker analysis of different intensity exercise groups
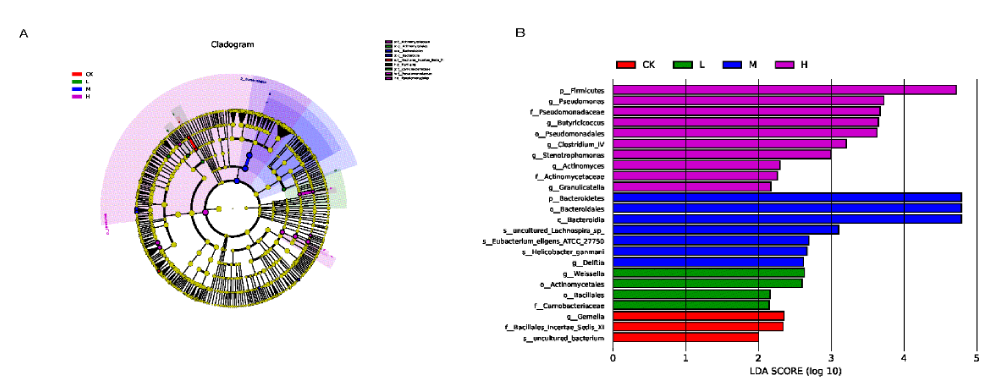
Find significant and different species between groups by Lefse analysis. Results of the analysis at the five levels of phylum, class, order, family and genus were presented by LDA score and LEfSe clade evolution dendrogram in Figure 4.
Figure 4: Biomarker analysis of different intensity exercise groups. A: LEfSe-species clade evolution of effect of exercise intensity on gut microbiota; B: LEfSe-LDA analysis of the effect of exercise intensity on gut microbiota./p>
LDA score and the nodes with different colors in the ring dendrogram (the light-yellow nodes represent the microbial taxa that were not significantly different in any of the different groups or between the groups), representing the microbial taxa that were significantly enriched in the corresponding groups and differences between the groups. After selecting the results with LDA score > 2, we found that there were 3 taxa in the CK group, 4 taxa in the L group, 7 taxa in the M group, and 10 taxa in the H group. p__Firmicutes, p__Bacteroidetes, g__Weissella, g__Gemella were abundant respectively in the H, M, L and CK groups.
Functional analysis
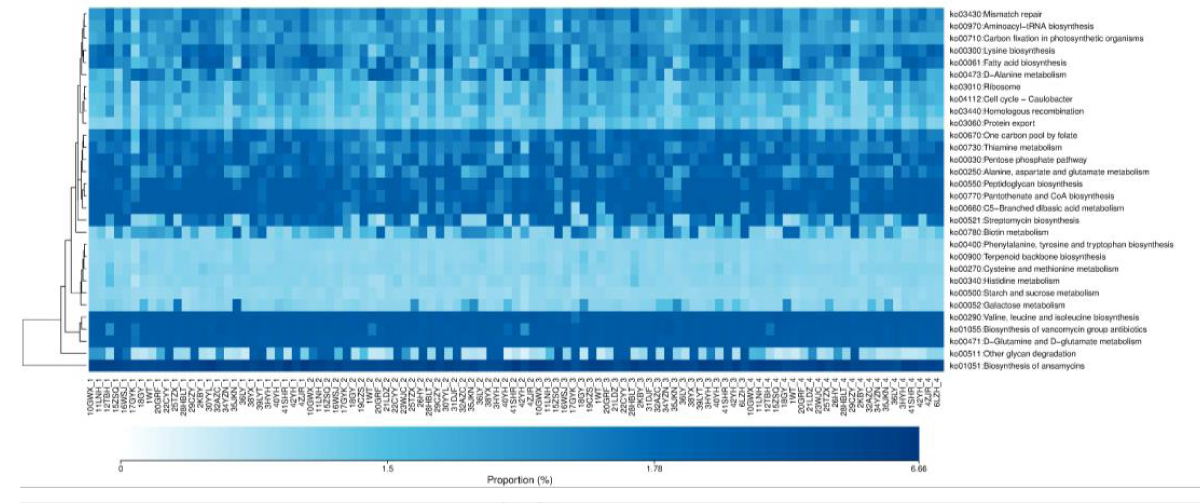
A total of 30 abundant metabolic pathways were identified in the four groups, of which 15 pathways were concentrated among the groups. The more common pathways are amino acid metabolism, terpenoid backbone biosynthesis, carbohydrate metabolism, cofactor and vitamin metabolism, lipid metabolism, cell growth and death, replication and repair of genetic information, etc. Most of these pathways are related to energy metabolism, growth, and development (Figure 5).
Figure 5: KEGG functional analysis of gut microbiota in athletes of different intensities. 1: CK; 2: L; 3: M; 4: H.
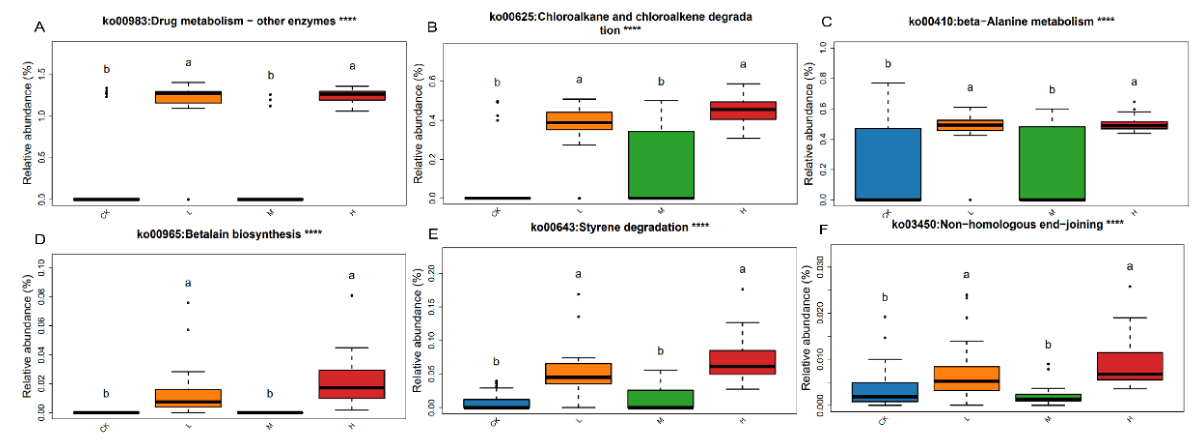
Annotation and enrichment analysis of KEGG biological pathways for the set of highly expressed genes with different exercise intensities, a total of 38 pathways were found to be significantly different (p < 0.05). With the increase of exercise intensity, compared with CK, ko00983 (biodegradation and metabolism of xenobiotics), ko00625 (Chloroalkane and chloroalkene degradation), ko00643 (Styrene degradation), ko00965 (Betalain biosynthesis), ko00410 (beta-Alanine metabolism) and ko03450 (non-homologous end-joining) were higher in the gut microbiota of athletes in L, M, and H. In the KEGG pathway, the microbial gene functions associated with xenobiotics biodegradation and metabolism, biosynthesis of secondary metabolites, amino acid metabolism, genetic information processing and lipid metabolism were also higher in the gut microbiome of the H group athletes (Figure 6).
Figure 6: Differential analysis of the KEGG pathway in the gut microbiota of pre- and post-exercise athletes. (A) Ko00983; (B) ko00625; (C) ko00410; (D) ko00965; (E) ko00643; (F) ko03450. Data were analyzed by one-way ANOVA, ****p < 0.0001./p>
Exercise has been found to have a beneficial impact on preventing and treating obesity, and alleviating symptoms of depression, anxiety, and cardiovascular disorders [16]. In addition, exercise is thought to influence, or correlate with differences in the gut microbiome across animal and human studies [17]. Hanna Dziewiecka, et al. [18] found that in non-athletes, increased physical activity significantly affected the relative abundance of short-chain fatty acids (SCFA). Sustained aerobic training for 60 minutes and physical activity at 60% HRmax or higher also affected the beta diversity index. Catarina Ramos [19] found that exercise/physical activity can beneficially alter the composition of the gut microbiome in young adults. However, research has yet to determine how changes in exercise intensity affect the gut microbiome. This study aims to elucidate how the gut microbiome differs among athletes who exercise at different intensities.
Diversity is important to promote stability and performance in all ecosystems [10]. Microbial diversity may become a novel indicator or biomarker of human health [20]. Loss of biodiversity in the gut microbiota is associated with several diseases such as obesity, metabolic diseases, inflammatory bowel diseases, or recurrent Clostridium difficile-associated diarrhea [21-23]. Exercise has been reported to be an important factor in increasing gut microbial diversity in humans [10]. In this study, we found that the α-diversity increased with exercise intensities but the correlation is not statistically significant. Recent studies revealed no increase in the abundance of gut microbiota in animal models with spontaneous movement. A possible reason is that the diversity of gut microbiota changes little in athletes who have long adapted to a certain sports training mode and diet structure.
In this study, most of the bacterial taxa in the gut microbiota of basketball players that responded to exercise intensity were bacteria of the Firmicutes. With the increase in the exercise intensity, the beneficial bacteria in the gut microbiota of the players increased and the enrichment of pathogenic bacteria decreased relatively. For example, the enrichment of Butyricoccus, Anaerostipes, Oxalobacter, and Clostridium_IV increased while Pseudomonas, Stenotrophomonas, Gemella, and Ruminococcus2 decreased relatively.
Butyricoccus and Anaerostipes are common high-abundance bacterial groups in the human intestine and both can metabolize and produce butyric acid. Butyrate has several beneficial properties that are essential to maintain gastrointestinal health. Therefore, butyrate-producing bacteria are seen as the next generation of probiotics. Butyrate is the preferred energy source for normal colonocytes, contributing to their normal homeostasis. It is a strong anti-tumor compound for tumor colonocytes, downregulating cell multiplication pathways, and promoting pro-apoptotic routes. The importance of probiotics in the prevention and treatment of CRC has been studied [24] and there is increasing evidence of the clinical significance of butyrate-producing gut microbes [25,26]. Wang, et al [27] reported that the four-carbon molecule butyrate may lower the risk of CRC in the absence of dysbiosis in the gut.
Oxalobacter is an anaerobic bacterium that colonizes the colon of a substantial proportion of the normal population and metabolizes dietary and endogenous oxalate. Therefore, oxalobacter can reduce the incidence of kidney stones. Studies begin to find a correlation between the proper balance of gut microbiota and a lower risk of oxalate kidney stones [28]. Oxalate homeostasis depends in part on the intestinal anaerobic bacterium Oxalobacter. A promising potential treatment is the intestinal elimination of endogenous oxalate using the two oxalate-degrading enzymes of Oxalobacter [29], an anaerobic microbe that normally inhabits the intestinal tract. Clostridium species, as a predominant cluster of commensal bacteria in our gut, exert lots of salutary effects on intestinal homeostasis [30]. Up to now, Clostridium species have been reported to attenuate inflammation and allergic diseases effectively owing to their distinctive biological activities [31]. Their cellular components and metabolites, like butyrate, secondary bile acids, and indole-propionic acid, play a probiotic role primarily through energizing intestinal epithelial cells, strengthening the intestinal barrier, and interacting with immune system [32]. In view of their salutary performances, Clostridium species have a huge potential as probiotics.
The professional athletes not only had distinct taxa composition characteristics but also had functional metabolism features in the gut microbiota [10,17]. In our study, further analysis of the functional prediction with the data obtained from the KEGG analysis indicated that the higher-level martial arts athletes had greater metabolic capacity in the gut microbiota than the lower-level athletes.
With the increase in exercise intensity, the levels of carbohydrate metabolism, amino acid metabolism, the metabolism of xenobiotics and glycans and metabolism in athletes were significantly improved, especially in group H. The main source of energy and nutrients available to the human gut microbiota is carbohydrates. Nutrients such as carbohydrates in food are fermented in the colon and cecum. SCFAs can be produced by anaerobic microorganisms in the large intestine [33]. Short-chain fatty acids can provide energy to the host and are important for host energy metabolism [34] and immune regulation [35,36]. The substances transformed or degraded by xenobiotics microorganisms are used as a source of energy, carbon, nitrogen, or other nutrients. Excessive release of H+ in skeletal muscle after high-intensity exercise causes acidosis, leading to a decrease in muscle pH and fatigue. However, β-alanine can delay the generation of fatigue and balance the overall pH [37]. Interestingly, in our study, the high-intensity athletes had a heavier exercise load than the low-intensity athletes. We also found that the beneficial bacteria obviously increased, improving the metabolic level of gut microbes in athletes.
However, as this is an exploratory study, we acknowledge the limitations of the data presented here, including the lack of an in-depth dietary analysis and a matched nonbasketball cohort. In future studies, we will recruit more basketball players and will include a healthy nonplayer cohort with both diet and exercise information, as diet and exercise are likely to play a role in influencing the taxonomic composition of the gut microbiota.
This preliminary study provides the first insight into the gut microbiota characteristics of professional college basketball athletes. We found that with increasing exercise intensity, the metabolic levels of high-intensity basketball players relatively increased. In the case of basketball players with long-term exercise, temporary exercise at different intensities cannot make a large impact on the diversity in their bodies. Maintaining a high-intensity basketball exercise can adjust the structure of the gut microbiota by increasing the healthy bacteria and inhibiting the harmful bacteria and therefore protect the human body from damage caused by a high-intensity exercise by enriching some beneficial bacteria.
The authors acknowledge all the study participants from the college basketball players of Southwest Jiaotong University. Hong-Xian Deng designed and conducted the study, and wrote the main manuscript text, Huan-Huan Li collected samples and analyzed data, Cui-Juan Wang suggested data analyses methods and review text, Jiang Liu managed the college basketball project, Jiu-Quan Qiao designed different intensity basketball training program for college students and supervised the analysis data, Yan Tong supervised the implementation of research projects, reviewed and revised the text. All authors reviewed the manuscript.
Funding
This work was supported by the Fund of Science and Technology Program of Sichuan Province (2021YFSY0048), Fundamental Research Funds for the Central Universities for Medical and Engineering Cooperation Projects (2682022ZTPY044, 2682021ZTPY015).
Compliance with ethical standards: Informed consent all participants provided informed consent.
- Silverman MN, Deuster PA. Biological mechanisms underlying the role of physical fitness in health and resilience. Interface Focus. 2014 Oct 6;4(5):20140040. doi: 10.1098/rsfs.2014.0040. PMID: 25285199; PMCID: PMC4142018.
- Mitchell CM, Davy BM, Hulver MW, Neilson AP, Bennett BJ, Davy KP. Does Exercise Alter Gut Microbial Composition? A Systematic Review. Med Sci Sports Exerc. 2019 Jan;51(1):160-167. doi: 10.1249/MSS.0000000000001760. PMID: 30157109.
- Kudelka MR, Stowell SR, Cummings RD, Neish AS. Intestinal epithelial glycosylation in homeostasis and gut microbiota interactions in IBD. Nat Rev Gastroenterol Hepatol. 2020 Oct;17(10):597-617. doi: 10.1038/s41575-020-0331-7. Epub 2020 Jul 24. PMID: 32710014; PMCID: PMC8211394.
- Bressa C, Bailén-Andrino M, Pérez-Santiago J, González-Soltero R, Pérez M, Montalvo-Lominchar MG, Maté-Muñoz JL, Domínguez R, Moreno D, Larrosa M. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS One. 2017 Feb 10;12(2):e0171352. doi: 10.1371/journal.pone.0171352. PMID: 28187199; PMCID: PMC5302835.
- Mohr AE, Jäger R, Carpenter KC, Kerksick CM, Purpura M, Townsend JR, West NP, Black K, Gleeson M, Pyne DB, Wells SD, Arent SM, Kreider RB, Campbell BI, Bannock L, Scheiman J, Wissent CJ, Pane M, Kalman DS, Pugh JN, Ortega-Santos CP, Ter Haar JA, Arciero PJ, Antonio J. The athletic gut microbiota. J Int Soc Sports Nutr. 2020 May 12;17(1):24. doi: 10.1186/s12970-020-00353-w. PMID: 32398103; PMCID: PMC7218537.
- Donati Zeppa S, Agostini D, Gervasi M, Annibalini G, Amatori S, Ferrini F, Sisti D, Piccoli G, Barbieri E, Sestili P, Stocchi V. Mutual Interactions among Exercise, Sport Supplements and Microbiota. Nutrients. 2019 Dec 20;12(1):17. doi: 10.3390/nu12010017. PMID: 31861755; PMCID: PMC7019274.
- Keohane DM, Woods T, O'Connor P, Underwood S, Cronin O, Whiston R, O'Sullivan O, Cotter P, Shanahan F, Molloy MGM. Four men in a boat: Ultra-endurance exercise alters the gut microbiome. J Sci Med Sport. 2019 Sep;22(9):1059-1064. doi: 10.1016/j.jsams.2019.04.004. Epub 2019 Apr 18. PMID: 31053425.
- Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. N Engl J Med. 2016 Dec 15;375(24):2369-2379. doi: 10.1056/NEJMra1600266. PMID: 27974040.
- Belcher BR, Zink J, Azad A, Campbell CE, Chakravartti SP, Herting MM. The Roles of Physical Activity, Exercise, and Fitness in Promoting Resilience During Adolescence: Effects on Mental Well-Being and Brain Development. Biol Psychiatry Cogn Neurosci Neuroimaging. 2021 Feb;6(2):225-237. doi: 10.1016/j.bpsc.2020.08.005. Epub 2020 Aug 18. PMID: 33067166; PMCID: PMC7878276.
- Clarke SF, Murphy EF, O'Sullivan O, Lucey AJ, Humphreys M, Hogan A, Hayes P, O'Reilly M, Jeffery IB, Wood-Martin R, Kerins DM, Quigley E, Ross RP, O'Toole PW, Molloy MG, Falvey E, Shanahan F, Cotter PD. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014 Dec;63(12):1913-20. doi: 10.1136/gutjnl-2013-306541. Epub 2014 Jun 9. PMID: 25021423.
- Codella R, Luzi L, Terruzzi I. Exercise has the guts: How physical activity may positively modulate gut microbiota in chronic and immune-based diseases. Dig Liver Dis. 2018 Apr;50(4):331-341. doi: 10.1016/j.dld.2017.11.016. Epub 2017 Nov 28. PMID: 29233686.
- Fugmann M, Breier M, Rottenkolber M, Banning F, Ferrari U, Sacco V, Grallert H, Parhofer KG, Seissler J, Clavel T, Lechner A. The stool microbiota of insulin resistant women with recent gestational diabetes, a high risk group for type 2 diabetes. Sci Rep. 2015 Aug 17;5:13212. doi: 10.1038/srep13212. PMID: 26279179; PMCID: PMC4538691.
- Mejía-León ME, Petrosino JF, Ajami NJ, Domínguez-Bello MG, de la Barca AM. Fecal microbiota imbalance in Mexican children with type 1 diabetes. Sci Rep. 2014 Jan 22;4:3814. doi: 10.1038/srep03814. PMID: 24448554; PMCID: PMC3898044.
- Li C, Huang Q, Yang R, Dai Y, Zeng Y, Tao L, Li X, Zeng J, Wang Q. Gut microbiota composition and bone mineral loss-epidemiologic evidence from individuals in Wuhan, China. Osteoporos Int. 2019 May;30(5):1003-1013. doi: 10.1007/s00198-019-04855-5. Epub 2019 Jan 21. PMID: 30666372.
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011 Jun 24;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. PMID: 21702898; PMCID: PMC3218848.
- Rawson RA, Chudzynski J, Gonzales R, Mooney L, Dickerson D, Ang A, Dolezal B, Cooper CB. The Impact of Exercise On Depression and Anxiety Symptoms Among Abstinent Methamphetamine-Dependent Individuals in A Residential Treatment Setting. J Subst Abuse Treat. 2015 Oct;57:36-40. doi: 10.1016/j.jsat.2015.04.007. Epub 2015 Apr 15. PMID: 25934458; PMCID: PMC4560957.
- Barton W, Penney NC, Cronin O, Garcia-Perez I, Molloy MG, Holmes E, Shanahan F, Cotter PD, O'Sullivan O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut. 2018 Apr;67(4):625-633. doi: 10.1136/gutjnl-2016-313627. Epub 2017 Mar 30. PMID: 28360096.
- Dziewiecka H, Buttar HS, Kasperska A, Ostapiuk-Karolczuk J, Domagalska M, Cichoń J, Skarpańska-Stejnborn A. Physical activity induced alterations of gut microbiota in humans: a systematic review. BMC Sports Sci Med Rehabil. 2022 Jul 7;14(1):122. doi: 10.1186/s13102-022-00513-2. PMID: 35799284; PMCID: PMC9264679.
- Ramos C, Gibson GR, Walton GE, Magistro D, Kinnear W, Hunter K. Systematic Review of the Effects of Exercise and Physical Activity on the Gut Microbiome of Older Adults. Nutrients. 2022 Feb 5;14(3):674. doi: 10.3390/nu14030674. PMID: 35277033; PMCID: PMC8837975.
- Shanahan F. Probiotics in perspective. Gastroenterology. 2010 Dec;139(6):1808-12. doi: 10.1053/j.gastro.2010.10.025. Epub 2010 Oct 19. PMID: 20965190.
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007 Jul;56(7):1761-72. doi: 10.2337/db06-1491. Epub 2007 Apr 24. PMID: 17456850.
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jørgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clément K, Doré J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T; MetaHIT consortium; Bork P, Wang J, Ehrlich SD, Pedersen O. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013 Aug 29;500(7464):541-6. doi: 10.1038/nature12506. PMID: 23985870.
- Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. 2015 Jan;37(1):47-55. doi: 10.1007/s00281-014-0454-4. Epub 2014 Nov 25. PMID: 25420450; PMCID: PMC4281375.
- Eslami M, Yousefi B, Kokhaei P, Hemati M, Nejad ZR, Arabkari V, Namdar A. Importance of probiotics in the prevention and treatment of colorectal cancer. J Cell Physiol. 2019 Aug;234(10):17127-17143. doi: 10.1002/jcp.28473. Epub 2019 Mar 25. PMID: 30912128.
- Chang SC, Shen MH, Liu CY, Pu CM, Hu JM, Huang CJ. A gut butyrate-producing bacterium Butyricicoccus pullicaecorum regulates short-chain fatty acid transporter and receptor to reduce the progression of 1,2-dimethylhydrazine-associated colorectal cancer. Oncol Lett. 2020 Dec;20(6):327. doi: 10.3892/ol.2020.12190. Epub 2020 Oct 5. PMID: 33101496; PMCID: PMC7577080.
- Gui Q, Li H, Wang A, Zhao X, Tan Z, Chen L, Xu K, Xiao C. The association between gut butyrate-producing bacteria and non-small-cell lung cancer. J Clin Lab Anal. 2020 Aug;34(8):e23318. doi: 10.1002/jcla.23318. Epub 2020 Mar 29. PMID: 32227387; PMCID: PMC7439349.
- Wang Y, Huang D, Chen KY, Cui M, Wang W, Huang X, Awadellah A, Li Q, Friedman A, Xin WW, Di Martino L, Cominelli F, Miron A, Chan R, Fox JG, Xu Y, Shen X, Kalady MF, Markowitz S, Maillard I, Lowe JB, Xin W, Zhou L. Fucosylation Deficiency in Mice Leads to Colitis and Adenocarcinoma. Gastroenterology. 2017 Jan;152(1):193-205.e10. doi: 10.1053/j.gastro.2016.09.004. Epub 2016 Sep 14. PMID: 27639802; PMCID: PMC5164974.
- Chamberlain CA, Hatch M, Garrett TJ. Metabolomic and lipidomic characterization of Oxalobacter formigenes strains HC1 and OxWR by UHPLC-HRMS. Anal Bioanal Chem. 2019 Jul;411(19):4807-4818. doi: 10.1007/s00216-019-01639-y. Epub 2019 Feb 11. PMID: 30740635; PMCID: PMC6612311.
- Hoppe B, Leumann E, Milliner DS. CHAPTER 33 - Urolithiasis and Nephrocalcinosis in Childhood. In Comprehensive Pediatric Nephrology eds. Geary DF, Schaefer. Philadelphia: Mosby. 2008; 499-525.
- Guo P, Zhang K, Ma X, He P. Clostridium species as probiotics: potentials and challenges. J Anim Sci Biotechnol. 2020 Feb 20;11:24. doi: 10.1186/s40104-019-0402-1. PMID: 32099648; PMCID: PMC7031906.
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013 Aug 8;500(7461):232-6. doi: 10.1038/nature12331. Epub 2013 Jul 10. PMID: 23842501.
- Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front Microbiol. 2016 Jun 28;7:979. doi: 10.3389/fmicb.2016.00979. PMID: 27446020; PMCID: PMC4923077.
- Wang SZ, Yu YJ, Adeli K. Role of Gut Microbiota in Neuroendocrine Regulation of Carbohydrate and Lipid Metabolism via the Microbiota-Gut-Brain-Liver Axis. Microorganisms. 2020 Apr 7;8(4):527. doi: 10.3390/microorganisms8040527. PMID: 32272588; PMCID: PMC7232453.
- Lyu Z, Wang L, Wang J, Wang Z, Zhang S, Wang J, Cheng J, Lai C. Oat bran and wheat bran impact net energy by shaping microbial communities and fermentation products in pigs fed diets with or without xylanase. J Anim Sci Biotechnol. 2020 Oct 8;11:99. doi: 10.1186/s40104-020-00505-7. PMID: 33062263; PMCID: PMC7542896.
- Reynolds LA, Finlay BB. A case for antibiotic perturbation of the microbiota leading to allergy development. Expert Rev Clin Immunol. 2013 Nov;9(11):1019-30. doi: 10.1586/1744666X.2013.851603. PMID: 24168410.
- Bai Y, Mansell TJ. Production and Sensing of Butyrate in a Probiotic Escherichia coli Strain. Int J Mol Sci. 2020 May 20;21(10):3615. doi: 10.3390/ijms21103615. PMID: 32443851; PMCID: PMC7279287.
- Artioli GG, Gualano B, Smith A, Stout J, Lancha AH Jr. Role of beta-alanine supplementation on muscle carnosine and exercise performance. Med Sci Sports Exerc. 2010 Jun;42(6):1162-73. doi: 10.1249/MSS.0b013e3181c74e38. PMID: 20479615.